Confident phosphorylation site localization using the Mascot Delta Score
06-Nov-2010
Molecular & Cellular Proteomics, 2010, doi: 10.1074/mcp.M110.003830, published on 06.11.2010
Molecular & Cellular Proteomics, online article
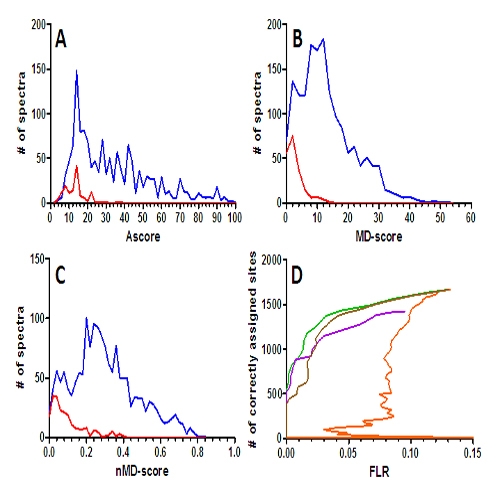
Large scale phosphorylation analysis is more and more getting into focus of proteomic research. While it is now possible to identify thousands of phosphorylated peptides in a biological system, confident site localization remains challenging. Here we validate the Mascot Delta Score (MD score) as a simple method that achieves similar sensitivity and specificity for phosphosite localization as the published Ascore which is mainly used in conjunction with Sequest. The MD-score was evaluated using LC-MS/MS data of 180 individually synthesised phosphopeptides with precisely known phosphorylation sites. We tested the MD-score for a wide range of commonly available fragmentation methods and found it to be applicable throughout with high statistical significance. However, the different fragmentation techniques differ strongly in their ability to localise phosphorylation sites. At 1% FLR, the highest number of correctly assigned phosphopeptides was achieved by higher energy collision induced dissociation (HCD) in combination with an Orbitrap mass analyzer followed very closely by low resolution ion trap spectra obtained after electron transfer dissociation (ETD). Both these methods are significantly better than low resolution spectra acquired after collision induced dissociation (CID) and multi stage activation (MSA). Score thresholds determined from simple calibration functions for each fragmentation method were stable over replicate analyses of the phosphopeptide set. The MD-score outperforms the Ascore for tyrosine phosphorylated peptides and we further show that the ability to call sites correctly increases with increasing distance of two candidate sites within a peptide sequence. The MD-score does not require complex computational steps which makes it attractive in terms of practical utility. We provide all mass spectra and the synthetic peptides to the community so that the development of present and future localization software can be benchmarked and any laboratory can determine MD-scores and localization probabilities for their individual analytical set up.