Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction
22-May-2008
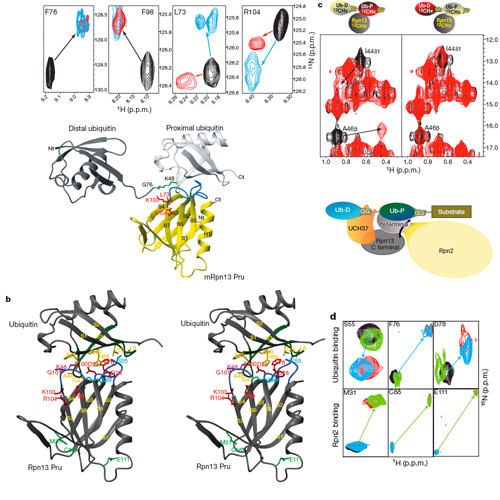
Targeted protein degradation is largely performed by the ubiquitin–proteasome pathway, in which substrate proteins are marked by covalently attached ubiquitin chains that mediate recognition by the proteasome. It is currently unclear how the proteasome recognizes its substrates, as the only established ubiquitin receptor intrinsic to the proteasome is Rpn10/S5a (ref. 1), which is not essential for ubiquitin-mediated protein degradation in budding yeast2. In the accompanying manuscript we report that Rpn13 (refs 3–7), a component of the nine-subunit proteasome base, functions as a ubiquitin receptor8, complementing its known role in docking de-ubiquitinating enzyme Uch37/UCHL5 (refs 4–6) to the proteasome. Here we merge crystallography and NMRdata to describe the ubiquitin-binding mechanism of Rpn13. We determine the structure of Rpn13 alone and complexed with ubiquitin. The co-complex reveals a novel ubiquitin-binding mode in which loops rather than secondary structural elements are used to capture ubiquitin. Further support for the role of Rpn13 as a proteasomal ubiquitin receptor is demonstrated by its ability to bind ubiquitin and proteasome subunit Rpn2/S1 simultaneously. Finally, we provide a model structure of Rpn13 complexed to diubiquitin, which provides insights into how Rpn13 as a ubiquitin receptor is coupled to substrate deubiquitination by Uch37. News articles: FAZ TUM Innovations Report IDW Report Internetchemie