Maintenance of imprinting and nuclear architecture in cycling cells
18-Sep-2007
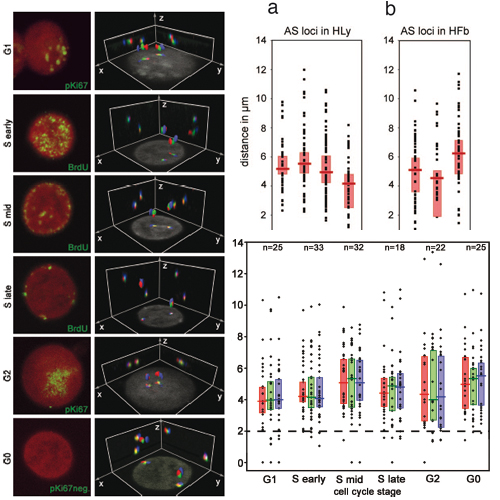
Dynamic gene repositioning has emerged as an additional level of epigenetic gene regulation. An early example was the report of a transient, spatial convergence (<2 micro m) of oppositely imprinted regions (‘‘kissing’’), including the Angelman syndrome/Prader– Willi syndrome (AS/PWS) locus and the Beckwith–Wiedemann syndrome locus in human lymphocytes during late S phase. It was argued that kissing is required for maintaining opposite imprints in cycling cells. Employing 3D-FISH with a BAC contig covering the AS/PWS region, light optical, serial sectioning, and quantitative 3D-image analysis, we observed that both loci always retained a compact structure and did not form giant loops. Three-dimensional distances measured among various, homologous AS/PWS segments in 393 human lymphocytes, 132 human fibroblasts, and 129 lymphoblastoid cells from Gorilla gorilla revealed a wide range of distances at any stage of interphase and in G0. At late S phase, 4% of nuclei showed distances <2 micro m, 49% showed distances >6 micro m, and 18% even showed distances >8 micro m. A similar distance variability was found for Homo sapiens (HSA) 15 centromeres in a PWS patient with a deletion of the maternal AS/PWS locus and for the Beckwith–Wiedemann syndrome loci in human lymphocytes. A transient kiss during late S phase between loci widely separated at other stages of the cell cycle seems incompatible with known global constraints of chromatin movements in cycling cells. Further experiments suggest that the previously observed convergence of AS/PWS loci during late S phase was most likely a side effect of the convergence of nucleolus organizer region-bearing acrocentric human chromosomes, including HSA 15.